|
FASPAD: A Tool for the Detection of Linear Signaling Pathways in Protein Interaction Networks
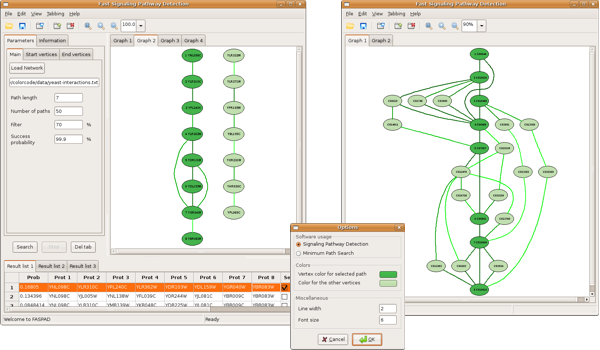
IntroductionFrom this page you can download a tool to detect signaling pathway candidates in protein interaction networks as described in
It can be also used to find minimum-weight simple paths of a user-specified length in arbitrary networks. The tool is based on an implementation of the algorithms described in the paper
LicenseThe program is distributed under the terms of the GNU Public License (GPL), version 2 or (at your option) any later version. See the file COPYING or http://www.gnu.org/copyleft/gpl.html for details. FASPAD uses the GraphViz library version 2.8 by AT&T Research Labs, which is distributed under the Common Public License (CPL). Therefore, as a special exception, you are allowed to link FASPAD with graphviz (or modified versions of graphviz that use the same license) and to distribute the resulting executable (providing you comply with the other terms of the GPL and CPL). Absolutely no guarantees or warranties are made concerning the suitability, correctness, or any other aspect of the distributed files. Any use is at your own risk. DownloadsLast updated on April 15, 2007.
As test data you can use the protein interaction networks from yeast and drosophila which both were obtained from http://bioinf.ucsd.edu/~sbandyop/GR/. AuthorsThe program was written by Falk Hüffner (homepage, email) and Thomas Zichner (email) in collaboration with Sebastian Wernicke (homepage, email). This work was supported by the Deutsche Telekom Stiftung and the Deutsche Forschungsgemeinschaft (DFG), projects PEAL (Parameterized Complexity and Exact Algorithms, NI 369/1), OPAL (Optimal Solutions for Hard Problems in Computational Biology, NI 369/2), and PIAF (Fixed-Parameter Algorithms, NI 369/4). EpilogueHappy pathway detecting! Thomas Zichner (email)
|